D
Drew Wanczowski
Guest
This is part three of our series about semantic RAG, in which we show you how to split, classify and vectorize long-form content. Be sure first to check out Part One: Enhancing GenAI with Multi-Model Data and Knowledge Graphs and Part Two: The Knowledge Graph.
Step two in our workflow is content preparation. Content preparation is a foundational process in Retrieval-Augmented Generation (RAG) that involves ingesting, chunking, vectorizing and classifying information to enrich the context provided to a language model.
We’ll start with ingesting the original content into Progress MarkLogic Server, preserving the source for lineage and user validation. Each record is then broken into smaller, manageable chunks to keep the content within the token limits of the model’s context window while enabling more precise targeting. These chunks are semantically classified using the knowledge graph we built in the previous article of our series, which adds valuable metadata like citational references and about-ness tags.
Finally, vector embeddings are applied to encode the meaning of each chunk numerically, allowing for similarity-based reranking. This allows only the most relevant content to be surfaced in response to user queries, enhancing the model’s ability to generate accurate and contextually grounded answers.
As part of this example, we will be preparing long-form content using the Progress MarkLogic Flux application. The MarkLogic Flux data movement tool is highly scalable and can be executed from the command line or embedded into your applications. The MarkLogic Flux tool will be used in conjunction with MarkLogic Server, the Semaphore platform and Azure OpenAI for embeddings. To build our data corpus, we will be using the PubMed Abstract content. Note the original files are in XML. The MarkLogic platform is multi-model in nature. We can store the XML alongside JSON and other data formats in a single database. We will keep the original XML intact for search and discovery, while the LLM will use data prepared in JSON format. Note that MarkLogic Flux supports many other data formats and can be adjusted for your workflow.
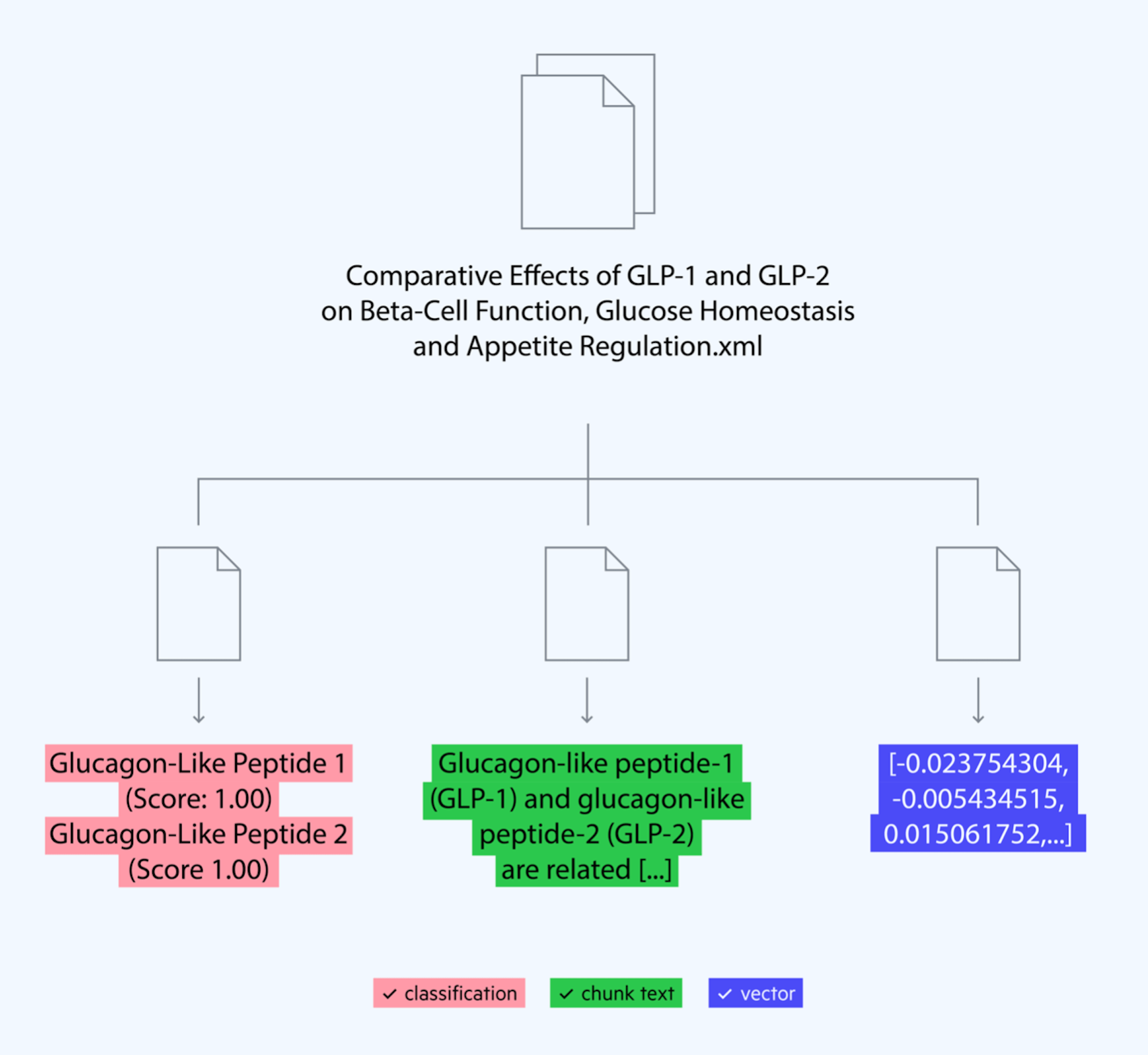
Image 1: Workflow showing you can store chunks, tags and embeddings in one document alongside the original content.
The complete Flux command will have options for loading, chunking, classifying and embedding generation. As we progress through our example, you will see the full set of options that you would use to ingest and process documents. One of the more common commands is import-files. We will scan a directory of XML files for this example.
Command Line:
First, we will accept an incoming file to be stored in MarkLogic Server. The content will be inserted as-is into the database. You can also run transformations of your choosing on these documents. Note that there are permissions and collections associated with this content. This means only people with these privileges can see the records. Additionally, collections allow for a logical grouping of content.
Next, we will be creating small chunks of data to be leveraged by the LLM. This is necessary because, while context windows—or the amount of information an LLM can process in a single prompt—continue to grow, they are still limited. This is why it’s important to be able to provide only the relevant information in a single prompt.
The process of “chunking” takes a larger body of content and breaks it into smaller amounts of data that can be inserted into the context window. These chunks can retain other metadata to support better data discovery. We will retain a pointer to the source file, semantic concepts from classification, vector embeddings and the original text. Let’s look at the chunking options provided by MarkLogic Flux.
This configuration will select a part of the article to be used for the textual content. In this case, we are looking at the abstract text of the article. You can also control the size of each chunk. For this, we will be setting the token limit on the higher side to contain the abstract text. Finally, we define the number of chunks we want per document and the output format. For our use case, we want one chunk per document. This will allow us to calculate lexical search relevance on each chunk. Relevance-based ranking of the chunks will be shown later in the article.
While chunks are good on their own, we would like to add additional metadata to enhance discoverability. We will use Semaphore Classification APIs to do semantic analysis of the text. This classification also utilizes a rich Natural Language Processing (NLP) engine and the knowledge model to determine key concepts in the text. The engine considers stemming and lemmatization of words and supporting evidence to assign concepts.
MarkLogic Server 12 supports native vector operations, including storing vector embeddings close to your data. Embeddings can be stored in the chunk record in JSON or XML formats. The LLMs let you create a vector representation of the content that can be used for similarity analysis. With Template Driven Extraction (TDE), we can specify a unique index for these embeddings to be used later as part of our discovery method.
MarkLogic Flux can directly call the Azure OpenAI embeddings model and other popular LLMs out of the box. Auto-wire the embedding model to include the utility:
A TDE can be generated to produce a view of the chunk metadata along with the vector embedding to be used in future queries.
Harnessing the power of TDEs, we can now create a graph of facts and information. This allows us to do multi-model queries against the corpus of data and helps visualize information for exploratory analysis or directly in an application’s UI.
This sample user interface builds a search application that allows you to review the articles and concepts. The semantic graph shows you the relation to the source file and the key concepts identified by classification. Note that the graph also shows parent terms, which can provide additional context.
When reviewing the answer from the LLM, you can come back to these documents to understand what content was used to derive the answer. This allows for full lineage and auditability of your GenAI application. The application end-user is always aware of the referenced content and how it fits within your organization.
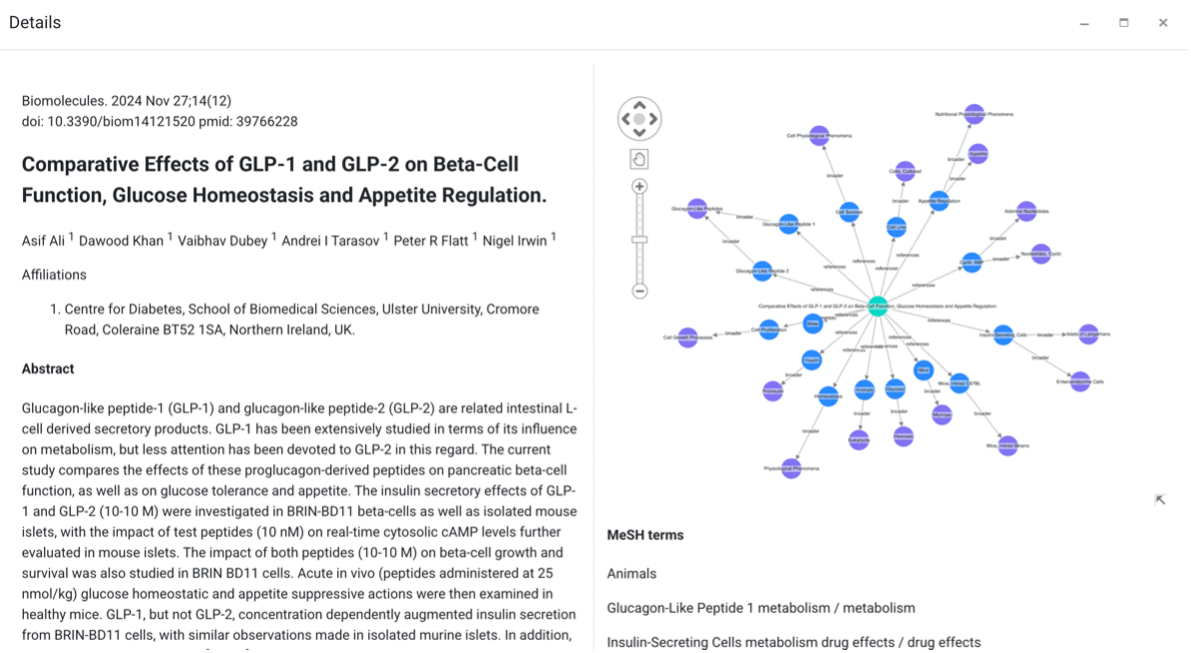
Image 2: Visualizing content and concepts of the knowledge graph. Content is directly linked to concepts that govern the domain.
In the next article, we’ll look at content discovery, how to orchestrate your hybrid query pipeline to maximize knowledge retrieval relevance and how to build a chat interface for your custom application.
To explore the full process for designing a RAG workflow to enhance the accuracy of your LLM responses, download our Semantic RAG whitepaper.
Continue reading...
Step two in our workflow is content preparation. Content preparation is a foundational process in Retrieval-Augmented Generation (RAG) that involves ingesting, chunking, vectorizing and classifying information to enrich the context provided to a language model.
We’ll start with ingesting the original content into Progress MarkLogic Server, preserving the source for lineage and user validation. Each record is then broken into smaller, manageable chunks to keep the content within the token limits of the model’s context window while enabling more precise targeting. These chunks are semantically classified using the knowledge graph we built in the previous article of our series, which adds valuable metadata like citational references and about-ness tags.
Finally, vector embeddings are applied to encode the meaning of each chunk numerically, allowing for similarity-based reranking. This allows only the most relevant content to be surfaced in response to user queries, enhancing the model’s ability to generate accurate and contextually grounded answers.
Preparing Long-Form Content
As part of this example, we will be preparing long-form content using the Progress MarkLogic Flux application. The MarkLogic Flux data movement tool is highly scalable and can be executed from the command line or embedded into your applications. The MarkLogic Flux tool will be used in conjunction with MarkLogic Server, the Semaphore platform and Azure OpenAI for embeddings. To build our data corpus, we will be using the PubMed Abstract content. Note the original files are in XML. The MarkLogic platform is multi-model in nature. We can store the XML alongside JSON and other data formats in a single database. We will keep the original XML intact for search and discovery, while the LLM will use data prepared in JSON format. Note that MarkLogic Flux supports many other data formats and can be adjusted for your workflow.

Image 1: Workflow showing you can store chunks, tags and embeddings in one document alongside the original content.
The complete Flux command will have options for loading, chunking, classifying and embedding generation. As we progress through our example, you will see the full set of options that you would use to ingest and process documents. One of the more common commands is import-files. We will scan a directory of XML files for this example.
Command Line:
./bin/flux import-files @options.txt First, we will accept an incoming file to be stored in MarkLogic Server. The content will be inserted as-is into the database. You can also run transformations of your choosing on these documents. Note that there are permissions and collections associated with this content. This means only people with these privileges can see the records. Additionally, collections allow for a logical grouping of content.
Code:
--path
/pubmed/
--connection-string
user:password@localhost:8000
--permissions
"rest-reader,read,rest-writer,update"
--collections
https://pubmed.ncbi.nlm.nih.govNext, we will be creating small chunks of data to be leveraged by the LLM. This is necessary because, while context windows—or the amount of information an LLM can process in a single prompt—continue to grow, they are still limited. This is why it’s important to be able to provide only the relevant information in a single prompt.
The process of “chunking” takes a larger body of content and breaks it into smaller amounts of data that can be inserted into the context window. These chunks can retain other metadata to support better data discovery. We will retain a pointer to the source file, semantic concepts from classification, vector embeddings and the original text. Let’s look at the chunking options provided by MarkLogic Flux.
Code:
--splitter-xpath
/PubmedArticle/MedlineCitation/Article/Abstract/AbstractText
--splitter-max-chunk-size
100000
--splitter-sidecar-max-chunks
1
--splitter-sidecar-document-type
JSON
--splitter-sidecar-root-name
chunk
--splitter-sidecar-collections
https://pubmed.ncbi.nlm.nih.gov/genai/chunksThis configuration will select a part of the article to be used for the textual content. In this case, we are looking at the abstract text of the article. You can also control the size of each chunk. For this, we will be setting the token limit on the higher side to contain the abstract text. Finally, we define the number of chunks we want per document and the output format. For our use case, we want one chunk per document. This will allow us to calculate lexical search relevance on each chunk. Relevance-based ranking of the chunks will be shown later in the article.
While chunks are good on their own, we would like to add additional metadata to enhance discoverability. We will use Semaphore Classification APIs to do semantic analysis of the text. This classification also utilizes a rich Natural Language Processing (NLP) engine and the knowledge model to determine key concepts in the text. The engine considers stemming and lemmatization of words and supporting evidence to assign concepts.
Code:
--classifier-host
localhost
--classifier-http
--classifier-port
5058MarkLogic Server 12 supports native vector operations, including storing vector embeddings close to your data. Embeddings can be stored in the chunk record in JSON or XML formats. The LLMs let you create a vector representation of the content that can be used for similarity analysis. With Template Driven Extraction (TDE), we can specify a unique index for these embeddings to be used later as part of our discovery method.
MarkLogic Flux can directly call the Azure OpenAI embeddings model and other popular LLMs out of the box. Auto-wire the embedding model to include the utility:
Code:
--embedder
azure
-Eapi-key=<your-api-key>
-Edeployment-name=text-test-embedding-ada-002
-Eendpoint=https://gpt-testing-custom-data1.openai.azure.comA TDE can be generated to produce a view of the chunk metadata along with the vector embedding to be used in future queries.
Code:
{
"template": {
"description": "Chunk View Template",
"context": "/chunk",
"collections": [ "https://pubmed.ncbi.nlm.nih.gov/genai/chunks" ],
"rows": [
{
"schemaName": "GenAI",
"viewName": "Chunks",
"viewLayout": "sparse",
"columns": [
{
"name": "source",
"scalarType": "string",
"val": "source-uri",
"nullable": false,
"invalidValues": "ignore"
},
{
"name": "text",
"scalarType": "string",
"val": "chunks/text",
"nullable": false,
"invalidValues": "ignore"
},
{
"name": "embedding",
"scalarType": "vector",
"nullable": true,
"val": "vec:vector(chunks/embedding)",
"dimension": "1536"
}
]
}
]
}
}Harnessing the power of TDEs, we can now create a graph of facts and information. This allows us to do multi-model queries against the corpus of data and helps visualize information for exploratory analysis or directly in an application’s UI.
Code:
{
"template": {
"description": "Content graph template",
"context": "/chunk",
"collections": [
"https://pubmed.ncbi.nlm.nih.gov/genai/chunks"
],
"templates": [
{
"context": "chunks/classification/META",
"triples": [
{
"subject": {
"val": "sem:iri(./ancestor::chunk/source-uri)",
"invalidValues": "ignore"
},
"predicate": {
"val": "sem:iri('http://purl.org/dc/terms/references')"
},
"object": {
"val": "xs:string(./id)"
}
}
]
}
]
}
}This sample user interface builds a search application that allows you to review the articles and concepts. The semantic graph shows you the relation to the source file and the key concepts identified by classification. Note that the graph also shows parent terms, which can provide additional context.
When reviewing the answer from the LLM, you can come back to these documents to understand what content was used to derive the answer. This allows for full lineage and auditability of your GenAI application. The application end-user is always aware of the referenced content and how it fits within your organization.

Image 2: Visualizing content and concepts of the knowledge graph. Content is directly linked to concepts that govern the domain.
In the next article, we’ll look at content discovery, how to orchestrate your hybrid query pipeline to maximize knowledge retrieval relevance and how to build a chat interface for your custom application.
To explore the full process for designing a RAG workflow to enhance the accuracy of your LLM responses, download our Semantic RAG whitepaper.
Continue reading...
